The glorious R::data.table
What is data.table?
data.table is an R package designed to make working with
data as easy and efficient as it can possibly be. At a first glance it
appears to update the standard R object data.frame. In case
you haven’t used them, you can still follow this tutorial. Basically
data.frames are R’s most flexible and broadly used way to
store data with a ‘spreadsheet’ structure, i.e., rows and columns.
But data.table adds a lot of functionality besides just
making data handling infinitely better. And it takes (and perfects)
ideas from all the best data handling tools:
- You can print tables on screen elegantly better than
tibble. - You can manipulate rows and columns and apply functions over them
(grouping by factors etc.) better than
dplyr. - You can effectively run loops better than the various
*plyfunctions. - You can chain commands like
magrittr(though …magrittrand native pipes are still great and work excellently in tandem withdata.table) - You can query tables with all the power of
SQL. - You can merge and join tables with the best of them, including very useful features like rolling joins.
- You can cast and melt tables better than
reshape2. - You can find “overlaps” between regions e.g. of genomic intervals,
like with
GRanges…
And because it does all this so elegantly, it is super easy to learn. To learn the most important stuff, let’s run an analysis on some Formula 1 racing data, because, of course.
Follow along in an Rstudio session. Make sure you run the commands, and most importantly, experiment with them.

Installation / loading
Reading and writing data
The data we’ll work with are from here. You can also find it on the Spartan storage server at /data/gpfs/projects/punim1869/shared_data/bioinfo_tutorials/f1.
Text-based table formats
Most text-based ways of storing data consist of lines of text (one
per row), and a separation character delimiting the column entries of
each row. For example, in the csv ([c]omma-[s]eparated
[v]alue) format,
"a","row","with",7,"columns","would","look","like","this".
Note that in X-separated formats, quote marks are often used to indicate
when a column contains text (c.f., say, numbers) because, to a computer,
numbers and the strings of text characters that represent them are very
different things.
The alternative to text-based data storage is binary files, which is a topic for another day.
fread()
data.table has its own table parsing function,
fread() ([f]ast [read]). It assumes you are giving it some
X-separated table and investigates the top of a the file (10,000 lines
by default) and tries to work out what the separator is and what the
kind of data in each column is, then reads it. Use it to read in the
data about lap times, drivers, and races—something like the
following:
The variables dr and lt now contain
data.table objects. Since these are enhanced
data.frames, we can do data.frame type things with them,
such as printing the contents by running the variable name alone:
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 841 20 3 1 1:32.713 92713
## ---
## 426631: 988 825 52 13 1:43.934 103934
## 426632: 988 825 53 13 1:44.164 104164
## 426633: 988 825 54 13 1:44.285 104285Notice it prints in a useful format showing the top and bottom few rows (default is five but you can play with it).
There are some useful arguments you can give to fread()
as necessary:
fread(header=F)- Often your tables won’t have a top row containing names. In this
case, tell
fread()not to assume the top row is column labels. It will call the columns “V1”, “V2”, … “Vn” by default, unless …
- Often your tables won’t have a top row containing names. In this
case, tell
fread(col.names=c("names","for","columns","go","in","this","vector"))- Obviously, for this to work the vector of column names must equal the number of columns.
fread(select=1:3)- This would read only the first three columns. With
select=c(6,3,5)we would get those columns in that order.
- This would read only the first three columns. With
Writing data
Is done with the normal R functions like write.csv and
saveRDS.
The central concept: Three indexing fields
A data.table is primarily manipulated using a
square-bracket syntax similar to data.frames. So you can,
for example, provide indices for rows and columns in the first two
arguments to select them, just like regular
data.frames:
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 5 1 1:32.342 92342## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 841 20 3 1 1:32.713 92713
## 4: 841 20 4 1 1:32.803 92803
## 5: 841 20 5 1 1:32.342 92342## raceId
## <int>
## 1: 841
## 2: 841
## 3: 841
## ---
## 426631: 988
## 426632: 988
## 426633: 988## lap
## <int>
## 1: 1
## 2: 2
## 3: 3
## ---
## 426631: 52
## 426632: 53
## 426633: 54## lap time
## <int> <char>
## 1: 1 1:38.109
## 2: 2 1:33.006
## 3: 3 1:32.713
## ---
## 426631: 52 1:43.934
## 426632: 53 1:44.164
## 426633: 54 1:44.285And you can always use the dollar sign operator $ to
extract a column as a vector …
## [1] "Lewis" "Nick" "Nico" "Fernando" "Heikki" "Kazuki"
## [7] "S̩bastien" "Kimi" "Robert" "Timo"## [1] "Lewis" "Nick" "Nico" "Fernando" "Heikki" "Kazuki"
## [7] "S̩bastien" "Kimi" "Robert" "Timo"If you want to select specific rows based on some property, you could use logical vectors (vectors of TRUEs and FALSEs that indicate whether a row should be selected):
## [1] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 841 1 1 2 1:40.573 100573
## ---
## 16193: 988 154 2 14 1:47.792 107792
## 16194: 988 825 1 20 2:00.490 120490
## 16195: 988 825 2 20 1:48.699 108699rm(selectMe) # It's good to clean up variables you don't need later to save memory
# Four steps ... really?… In reality, these operations are rarely used.
data.table’s natural method of doing things is almost
always better. Instead, we think of the indexing operator as having
three fields, that we use for different purposes. We will learn them in
order, but also pick up some other tricks on the way. To begin with,
memorise this:
dataTable[ <SELECT> , <DO_SOMETHING> , <GROUP_BY> ]
The [ <SELECT> ,,] field
While you’re working in the indexing fields, you can simply refer to column names of the table directly, and they will be treated as vectors. Which, as far as the computer’s understanding of a table goes, is exactly what they are.
The output of whatever you write in the SELECT field is used to (surprise!) select rows. Here are some typical examples:
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 841 1 1 2 1:40.573 100573
## ---
## 16193: 988 154 2 14 1:47.792 107792
## 16194: 988 825 1 20 2:00.490 120490
## 16195: 988 825 2 20 1:48.699 108699## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 842 20 1 1 1:49.614 109614
## ---
## 379: 987 20 2 1 2:18.392 138392
## 380: 988 20 1 3 1:47.471 107471
## 381: 988 20 2 3 1:43.632 103632This works because (say) lap<3 outputs a logical
vector that is TRUE when lap contains 1 or 2, and FALSE otherwise. But
what you output can also be numbers referring to rows. A classic example
is this trick used to sort the rows by some column …
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 977 1 69 4 1:07.411 67411
## 2: 977 1 65 4 1:07.424 67424
## 3: 977 817 69 3 1:07.442 67442
## ---
## 426631: 847 808 25 5 2:05:06.243 7506243
## 426632: 847 13 25 3 2:05:06.656 7506656
## 426633: 847 2 25 4 2:05:07.547 7507547Note the comma is optional but you should write it anyway for good reasons that you can ask me about.
Also note in Rstudio you can use <tab> to
autocomplete column names while you work inside the indexing fields.
Special variable: .N
Inside the indexing fields, .N always contains the
number of rows of the table (or the group; see later). Hence, to reverse
the table:
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 988 825 54 13 1:44.285 104285
## 2: 988 825 53 13 1:44.164 104164
## 3: 988 825 52 13 1:43.934 103934
## ---
## 426631: 841 20 3 1 1:32.713 92713
## 426632: 841 20 2 1 1:33.006 93006
## 426633: 841 20 1 1 1:38.109 98109%in% and %between%
These are exceptionally useful for selection:
## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 4 1 1:32.803 92803
## 2: 841 20 5 1 1:32.342 92342
## 3: 841 20 6 1 1:32.605 92605
## ---
## 31714: 988 825 5 20 1:47.197 107197
## 31715: 988 825 6 20 1:46.957 106957
## 31716: 988 825 7 19 1:46.344 106344## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 4 1 1:32.803 92803
## 2: 841 20 6 1 1:32.605 92605
## 3: 841 20 7 1 1:32.502 92502
## ---
## 23760: 988 825 4 20 1:46.760 106760
## 23761: 988 825 6 20 1:46.957 106957
## 23762: 988 825 7 19 1:46.344 106344## raceId driverId lap position time milliseconds
## <int> <int> <int> <int> <char> <int>
## 1: 841 20 1 1 1:38.109 98109
## 2: 841 20 2 1 1:33.006 93006
## 3: 841 20 3 1 1:32.713 92713
## ---
## 402869: 988 825 52 13 1:43.934 103934
## 402870: 988 825 53 13 1:44.164 104164
## 402871: 988 825 54 13 1:44.285 104285As you can imagine, this is something you might use to select, say, genes or SNPs falling within a particular range.
Merging
The SELECT field can also be used for one ofdata.table’s
most powerful functions, merging tables together. Notice in
lt that the drivers and races are given numbers, not
names.
If you look inside ra and dr, you will
notice these tables allow us to see what number corresponds to what
driver/race.
Merging allows us to use one data.table to look up what
rows of a second data.table it matches according to the
values in some column that occurs in both tables (say, driverId). When
you think about it, it is almost like the rows of one table are
selecting which rows of a second data table to join with. This
is why it makes sense to use the SELECT indexing field for merging.
To do it, we just put the selecting data.table in the
SELECT field of the other data.table, and then add an
argument listing which column(s) to look for matches on.
## driverId driverRef number code forename surname dob
## <int> <char> <int> <char> <char> <char> <char>
## 1: 20 vettel 5 VET Sebastian Vettel 03/07/1987
## 2: 20 vettel 5 VET Sebastian Vettel 03/07/1987
## 3: 20 vettel 5 VET Sebastian Vettel 03/07/1987
## ---
## 426631: 825 kevin_magnussen 20 MAG Kevin Magnussen 05/10/1992
## 426632: 825 kevin_magnussen 20 MAG Kevin Magnussen 05/10/1992
## 426633: 825 kevin_magnussen 20 MAG Kevin Magnussen 05/10/1992
## nationality url raceId lap
## <char> <char> <int> <int>
## 1: German http://en.wikipedia.org/wiki/Sebastian_Vettel 841 1
## 2: German http://en.wikipedia.org/wiki/Sebastian_Vettel 841 2
## 3: German http://en.wikipedia.org/wiki/Sebastian_Vettel 841 3
## ---
## 426631: Danish http://en.wikipedia.org/wiki/Kevin_Magnussen 988 52
## 426632: Danish http://en.wikipedia.org/wiki/Kevin_Magnussen 988 53
## 426633: Danish http://en.wikipedia.org/wiki/Kevin_Magnussen 988 54
## position time milliseconds
## <int> <char> <int>
## 1: 1 1:38.109 98109
## 2: 1 1:33.006 93006
## 3: 1 1:32.713 92713
## ---
## 426631: 13 1:43.934 103934
## 426632: 13 1:44.164 104164
## 426633: 13 1:44.285 104285Note data.table uses lists often so the dot character
. can be used as a shortcut for the list
function.
Also note you can join on more than one column, just add all the
names to the on= list.
For more flexibility, check the documentation with
?merge.data.table. You’ll find you can do cool things like
return rows that do NOT match. Let’s merge all our driver and race data
into the lap times table and move on.
If you look at lt now, you’ll see what happens when a
column name overlaps between the tables, (besides the one being merged
on; in this case, “url”). data.table automatically renames
one of them (e.g.) “i.url”.
The [, <DO_SOMETHING> ,] field
The DO_SOMETHING field is where the work gets done. Whether you are manipulating or adding or deleting columns, making calculations and returning the results, or even plotting, you do it all here. For example, you could do some calculation on the columns. Treat columns just like regular vectors. That’s how the computer thinks of them, so why shouldn’t you too?
## [1] 67411## [1] 1.123517# ... at an Australian Grand Prix (notice we are now also using the SELECT field)
lt[ name=="Australian Grand Prix" , min(milliseconds)/1000/60 ]## [1] 1.402083# ... and what years did Sebastian Vettel race in Australia?
lt[ name=="Australian Grand Prix" , unique(year) ]## [1] 2011 2012 2013 2014 2015 2016 1996 1997 1998 1999 2000 2001 2002 2003 2004
## [16] 2005 2006 2007 2008 2009 2010 2017# ... so what lap was Sebastian Vettel's fastest lap at the 2011 Australian Grand Prix?
lt[ name=="Australian Grand Prix" & year==2011 & surname=="Vettel" , lap[which(milliseconds==min(milliseconds))] ]## [1] 44Ceci n’est pas une pipe
This last line could be cleaner. If we need to filter multiple times, we can just chain indexing operations together. This can simplify the above command as follows:
lt[ name=="Australian Grand Prix" & year==2011 & surname=="Vettel" , ][ milliseconds==min(milliseconds), lap ]## [1] 44Command chaining like this makes data.table function as
a pipe! Just like magrittr! Brilliant!
I wonder how this lap compares to his other laps that day? Well, we could make a quick plot …
lt[ name=="Australian Grand Prix" & year==2011 & surname=="Vettel" , plot(x=lap,y=milliseconds/1000/60) ]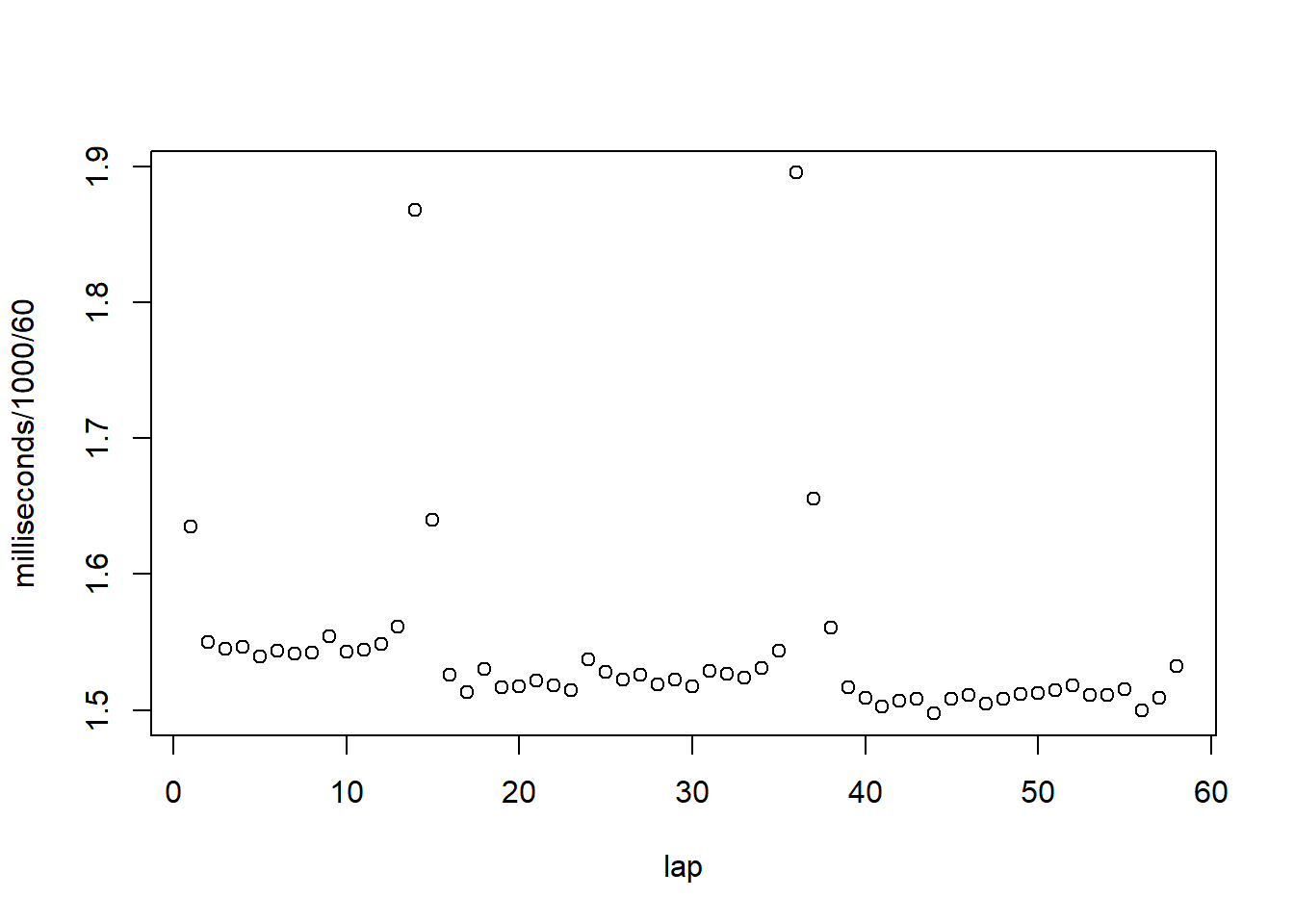
## NULL… looks like he had two pit stops and got faster and faster as the race went on. Which is good because .. he won! See?
lt[ name=="Australian Grand Prix" & year==2011 & surname=="Vettel", ][ lap==max(lap), paste("At the end of the final lap, Vettel was in position",position,"!") ]## [1] "At the end of the final lap, Vettel was in position 1 !"Returning a new data.table
Very often, the result you want is anotherdata.table. To
do this, we use the list( item1_name=<vector> , item2_name=<another vector> )
Each item in the list will become a column. Let’s use it to generate
some summary statistics for all the races together:
#Remember, `.()` is a shortcut for `list()`
lt[ , .( fastest_race_lap = min(milliseconds) , number_of_drivers = length(unique(driverId)) , average_driver_hours_per_race=sum(milliseconds)/3600000/length(unique(driverId)) ) ] ## fastest_race_lap number_of_drivers average_driver_hours_per_race
## <int> <int> <num>
## 1: 67411 123 92.3044Note, you can clean this code using multiple lines:
lt[ , .(
fastest_race_lap = min(milliseconds),
number_of_drivers = length(unique(driverId)),
average_driver_race_hours=sum(milliseconds)/3600000/length(unique(driverId))
)]A common use is to trim your data.table down to just
important columns. Let’s use it now to make a smaller table to look at
in future, one that only includes the Melbourne Grand Prix stats:
## [1] "Australian Grand Prix" "Malaysian Grand Prix"
## [3] "Chinese Grand Prix" "Turkish Grand Prix"
## [5] "Spanish Grand Prix" "Monaco Grand Prix"
## [7] "Canadian Grand Prix" "European Grand Prix"
## [9] "British Grand Prix" "German Grand Prix"
## [11] "Hungarian Grand Prix" "Belgian Grand Prix"
## [13] "Italian Grand Prix" "Singapore Grand Prix"
## [15] "Japanese Grand Prix" "Korean Grand Prix"
## [17] "Indian Grand Prix" "Abu Dhabi Grand Prix"
## [19] "Brazilian Grand Prix" "Bahrain Grand Prix"
## [21] "United States Grand Prix" "Austrian Grand Prix"
## [23] "Russian Grand Prix" "Mexican Grand Prix"
## [25] "Argentine Grand Prix" "San Marino Grand Prix"
## [27] "French Grand Prix" "Portuguese Grand Prix"
## [29] "Luxembourg Grand Prix" "Azerbaijan Grand Prix"Adding or modifying columns
The beautifully-named walrus operator := is
used to add, modify, or delete columns.
To add:
#add two columns using chaining
ltA[,seconds:=milliseconds/1000][,fullname:=paste(forename,surname)]
ltA## forename surname lap milliseconds position name
## <char> <char> <int> <int> <int> <char>
## 1: Sebastian Vettel 1 98109 1 Australian Grand Prix
## 2: Sebastian Vettel 2 93006 1 Australian Grand Prix
## 3: Sebastian Vettel 3 92713 1 Australian Grand Prix
## ---
## 19843: Daniel Ricciardo 23 90407 17 Australian Grand Prix
## 19844: Daniel Ricciardo 24 89466 17 Australian Grand Prix
## 19845: Daniel Ricciardo 25 89708 17 Australian Grand Prix
## circuitId year seconds fullname
## <int> <int> <num> <char>
## 1: 1 2011 98.109 Sebastian Vettel
## 2: 1 2011 93.006 Sebastian Vettel
## 3: 1 2011 92.713 Sebastian Vettel
## ---
## 19843: 1 2017 90.407 Daniel Ricciardo
## 19844: 1 2017 89.466 Daniel Ricciardo
## 19845: 1 2017 89.708 Daniel RicciardoTo modify:
## forename surname lap milliseconds position name
## <char> <char> <int> <int> <int> <char>
## 1: Sebastian Vettel 1 98109 1 Australian_Grand_Prix
## 2: Sebastian Vettel 2 93006 1 Australian_Grand_Prix
## 3: Sebastian Vettel 3 92713 1 Australian_Grand_Prix
## ---
## 19843: Daniel Ricciardo 23 90407 17 Australian_Grand_Prix
## 19844: Daniel Ricciardo 24 89466 17 Australian_Grand_Prix
## 19845: Daniel Ricciardo 25 89708 17 Australian_Grand_Prix
## circuitId year seconds fullname
## <int> <int> <num> <char>
## 1: 1 2011 98.109 Sebastian Vettel
## 2: 1 2011 93.006 Sebastian Vettel
## 3: 1 2011 92.713 Sebastian Vettel
## ---
## 19843: 1 2017 90.407 Daniel Ricciardo
## 19844: 1 2017 89.466 Daniel Ricciardo
## 19845: 1 2017 89.708 Daniel RicciardoTo destroy:
## forename surname lap position name circuitId year
## <char> <char> <int> <int> <char> <int> <int>
## 1: Sebastian Vettel 1 1 Australian_Grand_Prix 1 2011
## 2: Sebastian Vettel 2 1 Australian_Grand_Prix 1 2011
## 3: Sebastian Vettel 3 1 Australian_Grand_Prix 1 2011
## ---
## 19843: Daniel Ricciardo 23 17 Australian_Grand_Prix 1 2017
## 19844: Daniel Ricciardo 24 17 Australian_Grand_Prix 1 2017
## 19845: Daniel Ricciardo 25 17 Australian_Grand_Prix 1 2017
## seconds fullname
## <num> <char>
## 1: 98.109 Sebastian Vettel
## 2: 93.006 Sebastian Vettel
## 3: 92.713 Sebastian Vettel
## ---
## 19843: 90.407 Daniel Ricciardo
## 19844: 89.466 Daniel Ricciardo
## 19845: 89.708 Daniel RicciardoThe [,, <GROUP_BY> ] field
We can work out the average lap time over all these races (try this
yourself), but what about getting the average lap time for each
race? We need to perform something like mean(seconds), but
do it separately for each unique entry in the $year
column.
This is known as grouping, in this case, grouping by
$year. And we use the by= argument in the
## year V1
## <int> <num>
## 1: 2011 95.61877
## 2: 2012 98.46993
## 3: 2013 94.95944
## ---
## 20: 2009 97.42775
## 21: 2010 101.07681
## 22: 2017 91.10589Whoops! We never provided a column name so data.table
named it “V1”. Let’s fix that.
## year mean_laptime
## <int> <num>
## 1: 2011 95.61877
## 2: 2012 98.46993
## 3: 2013 94.95944
## ---
## 20: 2009 97.42775
## 21: 2010 101.07681
## 22: 2017 91.10589And if we want to know the lap time per year per driver, we can group on more variables …
## year fullname mean_laptime
## <int> <char> <num>
## 1: 2011 Sebastian Vettel 92.59067
## 2: 2011 Lewis Hamilton 92.97510
## 3: 2011 Mark Webber 93.24879
## ---
## 427: 2017 Marcus Ericsson 94.35571
## 428: 2017 Kevin Magnussen 93.98472
## 429: 2017 Daniel Ricciardo 98.88004… and perform multiple calculations …
## year fullname mean_laptime fastest_lap
## <int> <char> <num> <num>
## 1: 2011 Sebastian Vettel 92.59067 89.844
## 2: 2011 Lewis Hamilton 92.97510 90.314
## 3: 2011 Mark Webber 93.24879 89.600
## ---
## 427: 2017 Marcus Ericsson 94.35571 92.052
## 428: 2017 Kevin Magnussen 93.98472 87.568
## 429: 2017 Daniel Ricciardo 98.88004 89.447… and of course filter rows. Congratulations, we are now using all three indexing fields together!
ltA[ year > 2010 , .( mean_laptime = mean(seconds) , fastest_lap = min(seconds) ) , by=.(year,fullname) ]## year fullname mean_laptime fastest_lap
## <int> <char> <num> <num>
## 1: 2011 Sebastian Vettel 92.59067 89.844
## 2: 2011 Lewis Hamilton 92.97510 90.314
## 3: 2011 Mark Webber 93.24879 89.600
## ---
## 136: 2017 Marcus Ericsson 94.35571 92.052
## 137: 2017 Kevin Magnussen 93.98472 87.568
## 138: 2017 Daniel Ricciardo 98.88004 89.447This is a great example of how quickly data.table
aggregates good stats for plotting. We can very quickly use the above
data to look at how average lap times evolve over seasons, for example
…
## Loading required package: ggplot2ggplot(
data=ltA[ , .( mean_laptime = mean(seconds) , fastest_lap = min(seconds) ) , by=.(year,fullname) ],
mapping=aes(x=year,y=fastest_lap,colour=fullname)
) + geom_line() + theme(legend.position = "none")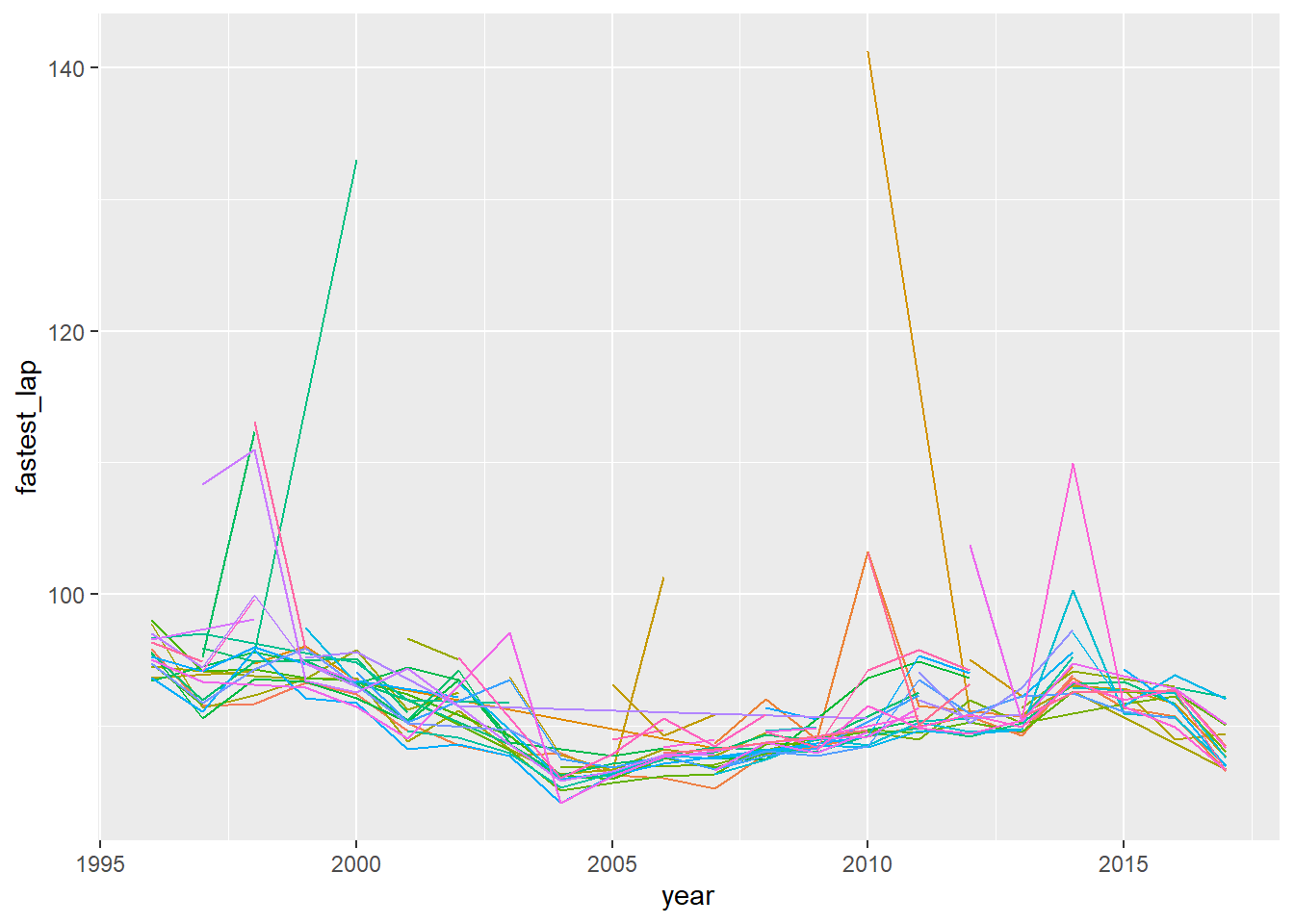
Getting slightly tricky
Initialise a new data.table from scratch
To make a data.table directly in R:
newDt <- data.table(
colCharacter = c("h","e","l","l","o","!"),
colInteger = 1L:2L,
colLogical = c(F,F,T)
)
newDt## colCharacter colInteger colLogical
## <char> <int> <lgcl>
## 1: h 1 FALSE
## 2: e 2 FALSE
## 3: l 1 TRUE
## 4: l 2 FALSE
## 5: o 1 FALSE
## 6: ! 2 TRUENotice the vectors you provide to fill the columns “recycle”, i.e.,
they repeat again and again until they match the length of the longest
vector. If they are not divisible evenly into that length
data.table will … actually that’s a good exercise, try it
and find out!
Group by with .N
Quite often you need to just count the members of each category, say, the number of laps each driver has driven. You could write …
## surname N
## <char> <int>
## 1: Vettel 430
## 2: Hamilton 580
## 3: Webber 525
## ---
## 101: Stroll 40
## 102: Vandoorne 55
## 103: Giovinazzi 55data.table offers this shortcut …
… which will automatically name the count column “N”
Special variable: .SD
Say you want to filter your data.table to select the
driver with the fastest lap in each race (or equivalently, filter BLAST
results, taking the highest-scoring alignment for each query … but this
is a strictly F1-based tutorial).
Naturally you will think about grouping by race. And about SELECTing a row based on lap time. But the SELECT field acts on the whole table, and you want to select from each group. What to do?
When grouping, the special variable .SD gives you access
to a mini data.table containing just the group it is working on.
## year forename surname lap position name circuitId
## <int> <char> <char> <int> <int> <char> <int>
## 1: 2011 Felipe Massa 55 9 Australian_Grand_Prix 1
## 2: 2012 Jenson Button 56 1 Australian_Grand_Prix 1
## 3: 2013 Kimi R\xcc_ikk̦nen 56 1 Australian_Grand_Prix 1
## ---
## 20: 2009 Nico Rosberg 48 7 Australian_Grand_Prix 1
## 21: 2010 Mark Webber 47 6 Australian_Grand_Prix 1
## 22: 2017 Kimi R\xcc_ikk̦nen 56 4 Australian_Grand_Prix 1
## seconds fullname
## <num> <char>
## 1: 88.947 Felipe Massa
## 2: 89.187 Jenson Button
## 3: 89.274 Kimi R\xcc_ikk̦nen
## ---
## 20: 87.706 Nico Rosberg
## 21: 88.358 Mark Webber
## 22: 86.538 Kimi R\xcc_ikk̦nensetorder() …
We’ve already seen one method of ordering a data.table,
something like: dt <- dt[order(varname),]. But this
requires copying the entire table, which is not memory efficient. To
sort the table more efficiently, we can use this function:
## forename surname lap position name circuitId year
## <char> <char> <int> <int> <char> <int> <int>
## 1: Sebastian Vettel 1 1 Australian_Grand_Prix 1 2011
## 2: Sebastian Vettel 2 1 Australian_Grand_Prix 1 2011
## 3: Sebastian Vettel 3 1 Australian_Grand_Prix 1 2011
## ---
## 19843: Daniel Ricciardo 23 17 Australian_Grand_Prix 1 2017
## 19844: Daniel Ricciardo 24 17 Australian_Grand_Prix 1 2017
## 19845: Daniel Ricciardo 25 17 Australian_Grand_Prix 1 2017
## seconds fullname
## <num> <char>
## 1: 98.109 Sebastian Vettel
## 2: 93.006 Sebastian Vettel
## 3: 92.713 Sebastian Vettel
## ---
## 19843: 90.407 Daniel Ricciardo
## 19844: 89.466 Daniel Ricciardo
## 19845: 89.708 Daniel Ricciardo## forename surname lap position name circuitId year
## <char> <char> <int> <int> <char> <int> <int>
## 1: Jean Alesi 1 5 Australian_Grand_Prix 1 1996
## 2: Jean Alesi 2 5 Australian_Grand_Prix 1 1996
## 3: Jean Alesi 3 5 Australian_Grand_Prix 1 1996
## ---
## 19843: Sebastian Vettel 55 1 Australian_Grand_Prix 1 2017
## 19844: Sebastian Vettel 56 1 Australian_Grand_Prix 1 2017
## 19845: Sebastian Vettel 57 1 Australian_Grand_Prix 1 2017
## seconds fullname
## <num> <char>
## 1: 106.506 Jean Alesi
## 2: 97.554 Jean Alesi
## 3: 96.830 Jean Alesi
## ---
## 19843: 87.369 Sebastian Vettel
## 19844: 87.437 Sebastian Vettel
## 19845: 88.709 Sebastian Vettel## forename surname lap position name circuitId year
## <char> <char> <int> <int> <char> <int> <int>
## 1: Jacques Villeneuve 1 1 Australian_Grand_Prix 1 1996
## 2: Jacques Villeneuve 2 1 Australian_Grand_Prix 1 1996
## 3: Jacques Villeneuve 3 1 Australian_Grand_Prix 1 1996
## ---
## 19843: Fernando Alonso 48 10 Australian_Grand_Prix 1 2017
## 19844: Fernando Alonso 49 10 Australian_Grand_Prix 1 2017
## 19845: Fernando Alonso 50 10 Australian_Grand_Prix 1 2017
## seconds fullname
## <num> <char>
## 1: 103.702 Jacques Villeneuve
## 2: 97.036 Jacques Villeneuve
## 3: 95.959 Jacques Villeneuve
## ---
## 19843: 90.077 Fernando Alonso
## 19844: 90.493 Fernando Alonso
## 19845: 92.485 Fernando Alonso… and more importantly, setkey()
There are computational advantages to having sorted tables. The
reason is the same reason dictionaries put the words in alphabetical
order and not randomly. When you search a dictionary for a word you
(probably without thinking about it) implement a kind of binary
search algorithm. data.table is often asked to find
values, in fact that’s most of that the SELECT field does. If
data.table knows that one or more columns are sorted, it
can search them far more quickly. setkey() does this:
setkey(ltA,forename,lap) #Note you cannot use `-` to reverse the order. That's why `setorder()` still exists
#This shows you that the data.table now has an attribute indicating which columns are sorted, or "keys", in the lingo
attributes(ltA)["sorted"]## $sorted
## [1] "forename" "lap"setorder(ltA,-year)
#Obviously, if you reorder the rows later the keys will be removed.
attributes(ltA)["sorted"]## $<NA>
## NULLThis becomes especially super important when performing very large merges, because these require a lot of searching behind the scenes to work out what rows match.
setnames()
This is a good way to change column names. The syntax is
setnames(dataTable,c("vector","of","old","names"),c("VECTOR","OF","NEW","NAMES"))
## [1] "colCharacter" "colInteger" "colLogical"## [1] "colChar" "colInt" "colLogical"# I like to use it in conjunction with magrittr pipes (if you don't get this command don't worry, magrittr can be a tutorial for another day)
require(magrittr)## Loading required package: magrittr## Warning: package 'magrittr' was built under R version 4.5.1## A B C
## <char> <int> <lgcl>
## 1: h 1 FALSE
## 2: e 2 FALSE
## 3: l 1 TRUE
## 4: l 2 FALSE
## 5: o 1 FALSE
## 6: ! 2 TRUEsetDT()
Similar to functions like as.matrix() and
as.data.frame(), there is an as.data.table()
function that will attempt to convert other objects into
data.tables. This requires a lot of copying of data behind
the scenes. Unless, of course, the object you are converting is a
data.frame because as we discussed, a
data.table is just an extension of a
data.frame. To really bring that home:
## [1] "data.table" "data.frame"So, why bother copying? setDT(dataFrame) will instantly
convert a dataFrame into a data.table with
almost zero extra memory or processing required.
This and the other “set” functions above all save memory using this this zero-copying paradigm, so now is a good time to talk about that more generally.
Pass by reference, the key to speed and memory efficiency
This is technical background, feel free to skip it. But as you get more advanced with R, it will become very important.
Normally, when you pass a function an object in R, the function makes a copy of the object and works on the copy (oversimplified but, in general that’s true). Copying data is inefficient, but can help programs to act “safely”, since you are less likely to end up accidentally modifying an object you don’t want to. This paradigm is known as copy on modify semantics.
data.table avoids making copies unless absolutely
necessary. This is partly why it is so much faster and uses less memory
than the same operations in tidyverse packages. It also bears some
risks. Consider this function that involves adding a column to a
data.table:
If we run this on newDt as defined above, nothing (well
… NULL) is returned. But look at the table before and after!
## [1] "A" "B" "C"## NULL## [1] "A" "B" "C" "newCol"As you can see, even though we never specifically over-wrote our
original table, it has been modified. that is because the walrus
operator function (:=), like many data.table
functions, uses reference semantics, that is, it is passed the
memory address of the actual original table, which it them modifies. So
you have to be careful. Other data.table operations will
indeed copy part of or the whole table, but always aims to optimise
speed and memory usage without being too risky. I mean … there’s no real
harm in having an extra column floating around, but imagine if we
deleted a column! That could actually be a problem. Be alert but not
alarmed.
Multiline DO_SOMETHING
DO_SOMETHING can get very complex. Use curly braces ({
and }) to denote multi-command operations in there. I
speculate that a driver’s fastest lap of a race is usually a few laps
before the last lap, when they are pushing the car hard but the tyres
have not yet worn out too much. Let’s check how many laps from the end
of the race drivers normally complete their fastest lap of the race.
fastLapDt <- lt[,{
lastLap <- max(lap)
verstappenFastest <- .SD[,lap[milliseconds==min(milliseconds)]]
.( lapsBeforeLastFastest = verstappenFastest - lastLap )
},by=.(circuitId,year)]
hist( fastLapDt$lapsBeforeLastFastest, breaks=function(x){min(x):0} )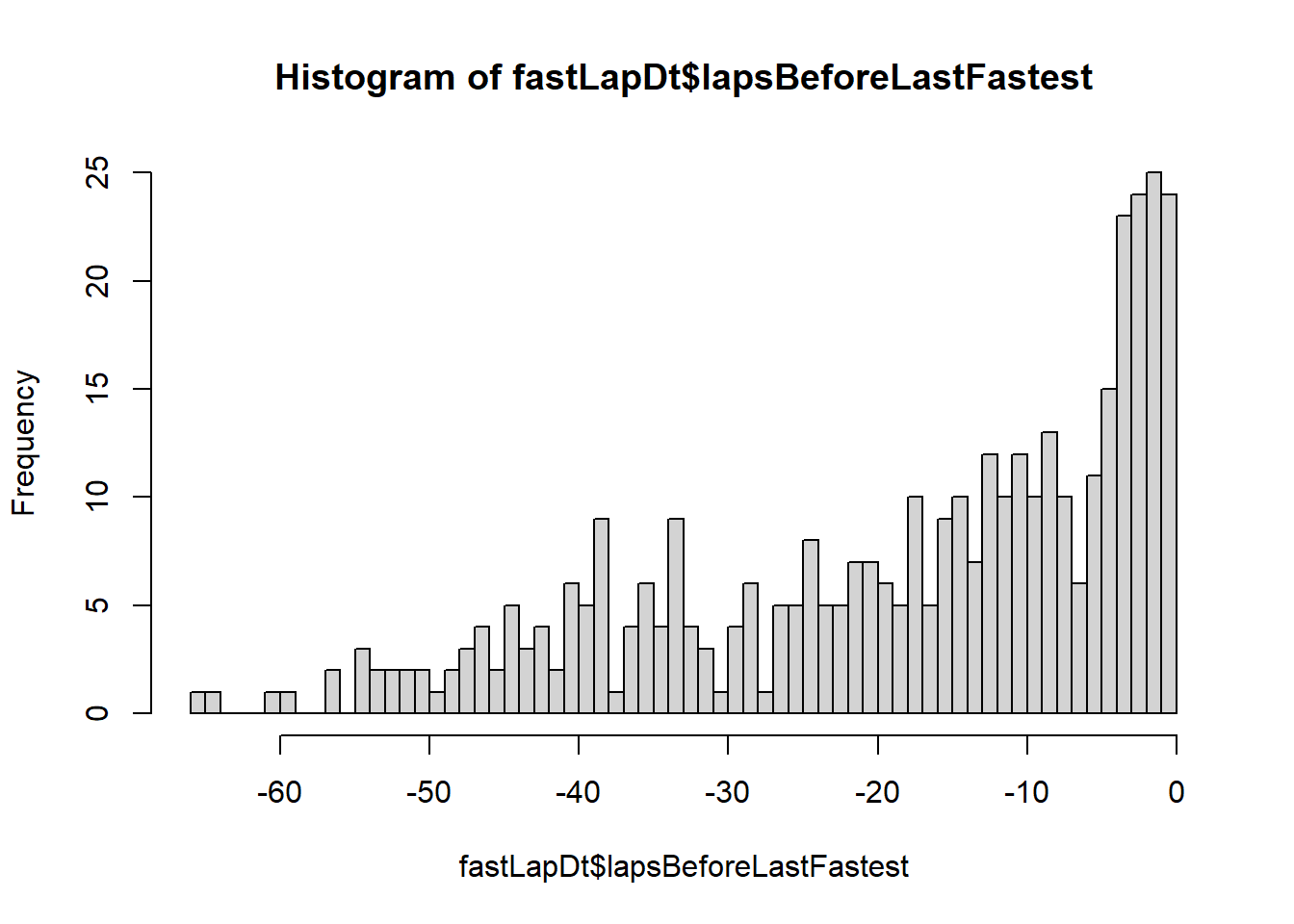
Yep, looks like it’s most often the second to last lap, but by a tiny margin.
Take some time to make sure you understand every part of this command, it contains many tricks and is a good recap of the tutorial so far.
Want a fun challenge? Figure out a way to do this with one line.
Use as a for loop
The GROUP_BY field effectively runs a set of commands for each group.
When you think about it, that’s quite similar to what a
for(){ ... } loop does. R for-loops are famously
slow. There’s a fascinating history as to why, it’s partially on
purpose to encourage you to use faster alternatives. *ply-type
functions are good, but using data.tables to mimic
for-loops, as we have done above, is even faster.
You can even modify a variable outside the data.table
while it is “looping”, using the <<-
operator.
Getting extra tricky
Special variable: .GRP
When you GROUP_BY, the groups are given a number you can access with the special variable .GRP. I can’t think of a use case right now but remember it, it does occasionally come in very handy.
Casting and melting
It’s very common to want to pivot a table from a wide shape to long shape or back. Intuitively, wide is like this:
| Driver | Lap 1 | Lap 2 | Lap 3 | Lap 4 |
|---|---|---|---|---|
| Driver 1 | 1:18.5 | 1:18.2 | 1:17.9 | 1:18.2 |
| Driver 2 | 1:17.2 | 1:17.0 | 1:16.5 | 1:17.2 |
| Driver 3 | 1:18.8 | 1:17.1 | 1:18.4 | 1:20.0 |
| Driver 4 | 1:19.1 | 1:17.0 | 1:15.9 | 1:20.2 |
… and long would be like this:
| Driver | Lap | Time |
|---|---|---|
| Driver 1 | Lap 1 | 1:18.5 |
| Driver 1 | Lap 2 | 1:18.2 |
| Driver 1 | Lap 3 | 1:17.9 |
| Driver 1 | Lap 4 | 1:18.2 |
| Driver 2 | Lap 1 | 1:17.2 |
| Driver 2 | Lap 2 | 1:17.0 |
| Driver 2 | Lap 3 | 1:16.5 |
| etc … | — | — |
Changing the shape of data from wide to long is called melting (picture melting cheese dripping from a pizza: it starts out widely distributed over the base and ends up as long vertical strings [columns]). Long –> wide is called casting, in a questionably successful attempt to maintain semantic symmetry with its counterpart.
Casting
Shape-shifting data requires some interesting syntax. Casting (going from long to wide) is (contrary to popular opinion) easier. The table you want is a kind of two-dimensional grid (look above), so you just specify the two variable whose levels should be represented on each axis of the grid. In this case you would specify that the drivers should be on the row axis, and the laps should be on the column axis.
Here’s a demo, using data.tables casting function
dcast().
## Driver lap seconds
## <char> <int> <num>
## 1: Antonio Giovinazzi 1 107.489
## 2: Antonio Giovinazzi 2 94.891
## 3: Antonio Giovinazzi 3 93.874
## ---
## 940: Valtteri Bottas 55 88.670
## 941: Valtteri Bottas 56 86.593
## 942: Valtteri Bottas 57 87.507## Using 'seconds' as value column. Use 'value.var' to override## Key: <Driver>
## Driver 1 2 3 4
## <char> <num> <num> <num> <num>
## 1: Antonio Giovinazzi 107.489 94.891 93.874 94.121
## 2: Carlos Sainz 101.891 91.386 91.658 90.646
## 3: Daniel Ricciardo 296.773 92.510 91.619 92.111
## ---
## 18: Sergio P̩rez 102.868 91.589 91.539 91.043
## 19: Stoffel Vandoorne 106.762 94.509 93.571 93.110
## 20: Valtteri Bottas 96.009 90.141 89.635 89.301heatmap(ltA17LapTimesCast[,-"Driver"] %>% as.matrix,Rowv = NA,Colv = NA,labRow = iconv(ltA17LapTimesCast$Driver))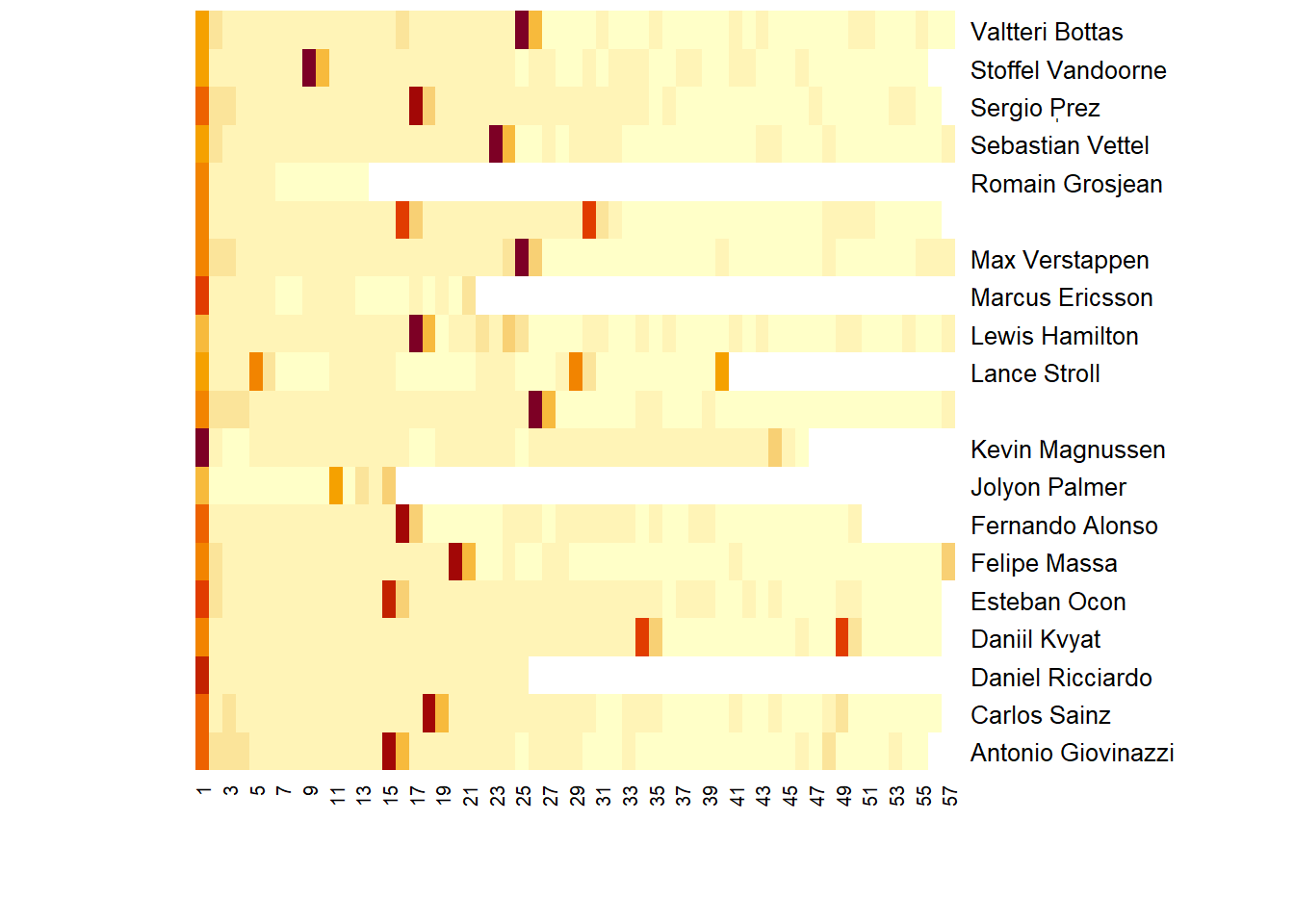
So that worked but we had a warning:
“Using ‘seconds’ as value column. Use ‘value.var’ to override”
This is for a very good reason. Casting creates a grid. We told the
function what variables (AKA columns) should define the rows and columns
of the grid. But we didn’t specify which column should be used to put
values in the actual grid itself. In this case we asked for a grid
defined by drivers and laps, there was only one column (“seconds”) left
over to provide values for the grid. But it ain’t necessarily
so. There will often be many columns to choose from, and you need to
specify which is the value-filling columns using the argument
value.var=....
A related case that comes up often is that there isn’t one unique (to
use this example) lap column for each driver, or vice versa. For
example, if there were two rows recording a time for the driver “Carlos
Sainz” in lap 4, then which one should provide the “seconds” to fill the
grid position corresponding to that driver/lap combination? One option
would be to, say, just take all the candidate rows and add their values
together. In this case, ‘adding’ would be a form of aggregation
— a way of compressing multiple values into one. Faced with multiple
values per grid item, data.table will ask you to provide a
function to use for aggregation.
Melt
Melt is (sort of) the opposite of cast. So we’ll use it to (sort of) recover the original table from the one we just produced. We can’t normally recover the exact table, because some information is normally lost in casting (for example, by aggregating multiple values into one). But in this instance we can!
If you look at those two tables above and think about how you would convert wide to long, you might notice there are some bits of information that need to be given.
- Which columns should provide data to be compressed into the new column? ( “1” , “2” , “3” , … )
- There will be a new column that tells you what original column the data came from. What should that be called? (“Lap”)
- What should the new column of data be called? (“Time”)
You provide this information in arguments to melt(),
using the arguments measure.vars (1),
value.name (2), and variable.name (3):
# We want to melt all the columns besides the first one, so let's make a list of their names ("1", "2", "3", ...)
meltVars <- colnames(ltA17LapTimesCast)[-1] # Do you know this trick? Cool, right?
melt(ltA17LapTimesCast,measure.vars=meltVars,value.name="Lap",variable.name="Time")## Driver Time Lap
## <char> <fctr> <num>
## 1: Antonio Giovinazzi 1 107.489
## 2: Carlos Sainz 1 101.891
## 3: Daniel Ricciardo 1 296.773
## ---
## 1138: Sergio P̩rez 57 NA
## 1139: Stoffel Vandoorne 57 NA
## 1140: Valtteri Bottas 57 87.507Casting and melting come up all the time in genetics. For example,
imagine having a list of gene expression values with a column for
‘treatment’. You’d likely want to turn that into a matrix and plot it,
so dcast would come into play. They are tricky operations,
but they make more sense once you have a use case of your own.
Helpful functions
These make everyday tasks easy and fast … in fact the prefix ‘f’ on a
lot of data.table functions stands for “fast”—they are
speed-optimised implementations of popular functions.
foverlaps
This function (“fast overlaps”, or I like to think of it as “find overlaps”) is made for genomics.
And is hence best understood with a genomics example. It solves the classic problem of needing to find the overlaps between genomic features, for example, when you want to know what genes overlap some genomic region, say, a QTL. In such a case you might have a data.table called “genes” describing gene positions, so it would have columns named something like “chromosome”, “start”, and “end”, and perhaps some kind of gene description called “description”. Let’s make one …
genes <- data.table(
chr = c("chr1","chr3","chr3","chr4"),
start = c( 1239, 120, 450, 8953),
end = c( 1278, 183, 620, 9232),
description = c("abc1","xyz2","def3","uvw4")
)And you would have a second data.table describing the QTL ranges:
QTLs <- data.table(
chr = c("chr3","chr4","chr5"),
start = c( 98, 1302, 8953),
end = c( 1034, 9032, 9232),
description = c("qtl1","qtl2","qtl3")
)So, in this case, qtl1 fully overlaps both genes on chr3, and qtl3 partially overlaps the gene on chr4.
Remember setkey() from earlier? foverlaps()
requires keyed data.tables (for very good speed-related reasons), and it
works by assuming the first key column is the chromosome, and
the next two are the start and end coordinates of the
feature. so we set up our data.tables accordingly:
That may look a bit silly because we conveniently named our columns the same in both data tables. But this function has many uses, and your columns won’t always have the same names. Imagine for example checking which festivals overlap with what season. Or what towns fall within certain ranges of latitudes. Or what experimental plots fall within certain distances of a water source. It’s flexible.
Let’s find the overlaps using foverlaps(), starting with
all instances where any part of a gene overlaps a QTL.
## Key: <chr, i.start, i.end>
## chr start end description i.start i.end i.description
## <char> <num> <num> <char> <num> <num> <char>
## 1: chr1 NA NA <NA> 1239 1278 abc1
## 2: chr3 98 1034 qtl1 120 183 xyz2
## 3: chr3 98 1034 qtl1 450 620 def3
## 4: chr4 1302 9032 qtl2 8953 9232 uvw4Great, right? Notice in the output the prefix ‘i.’ is prepended to columns names in the first data.table given to the function, so you can distinguish them from the columns in the other.
Remember in the section on merging how merges can be conceptualised as one data.table SELECTing rows from another? The same idea applies here: the first data.table selects rows from the second. And like merge, if a row in the selecting data.table that doesn’t find a match, it will produce a row with NAs in the relevant columns.
For example, let’s switch it around and ask the QTLs to find genes they overlap. The QTL on chr5 isn’t going to hit any genes, so notice we get NAs there:
## Key: <chr, i.start, i.end>
## chr start end description i.start i.end i.description
## <char> <num> <num> <char> <num> <num> <char>
## 1: chr3 120 183 xyz2 98 1034 qtl1
## 2: chr3 450 620 def3 98 1034 qtl1
## 3: chr4 8953 9232 uvw4 1302 9032 qtl2
## 4: chr5 NA NA <NA> 8953 9232 qtl3We can suppress output from unmatched rows using
nomatch=0.
## Key: <chr, i.start, i.end>
## chr start end description i.start i.end i.description
## <char> <num> <num> <char> <num> <num> <char>
## 1: chr3 120 183 xyz2 98 1034 qtl1
## 2: chr3 450 620 def3 98 1034 qtl1
## 3: chr4 8953 9232 uvw4 1302 9032 qtl2And finally, say we only want to select items where the (say) gene,
falls entirely within the (say) QTL, and doesn’t just overlap
some bit of it. Modify the type= argument.
## Key: <chr, i.start, i.end>
## chr start end description i.start i.end i.description
## <char> <num> <num> <char> <num> <num> <char>
## 1: chr3 98 1034 qtl1 120 183 xyz2
## 2: chr3 98 1034 qtl1 450 620 def3fifelse
Just like ifelse(), but faster …
dr[1:10,ifelse(
nationality=="German",
paste(forename,"wants leberwurst and saurkraut for breakfast"),
paste(forename,"doesn't want pulverised offal for breakfast")
)]## [1] "Lewis doesn't want pulverised offal for breakfast"
## [2] "Nick wants leberwurst and saurkraut for breakfast"
## [3] "Nico wants leberwurst and saurkraut for breakfast"
## [4] "Fernando doesn't want pulverised offal for breakfast"
## [5] "Heikki doesn't want pulverised offal for breakfast"
## [6] "Kazuki doesn't want pulverised offal for breakfast"
## [7] "S̩bastien doesn't want pulverised offal for breakfast"
## [8] "Kimi doesn't want pulverised offal for breakfast"
## [9] "Robert doesn't want pulverised offal for breakfast"
## [10] "Timo wants leberwurst and saurkraut for breakfast"frank
Super useful. Let’s say you want to plot the driver times on the first lap of Melbourne 2017, in order of speed. In other words, you want to rank them …
ltA27lap1 <- ltA[lap==1 & year==2017,.(
seconds,
rank=frank(seconds)
)]
plot(ltA27lap1$rank,ltA27lap1$seconds,pch=20) # That super-slow time is result of Daniel Ricciardo's fuel cell shitting itself during the formation lap. Boo.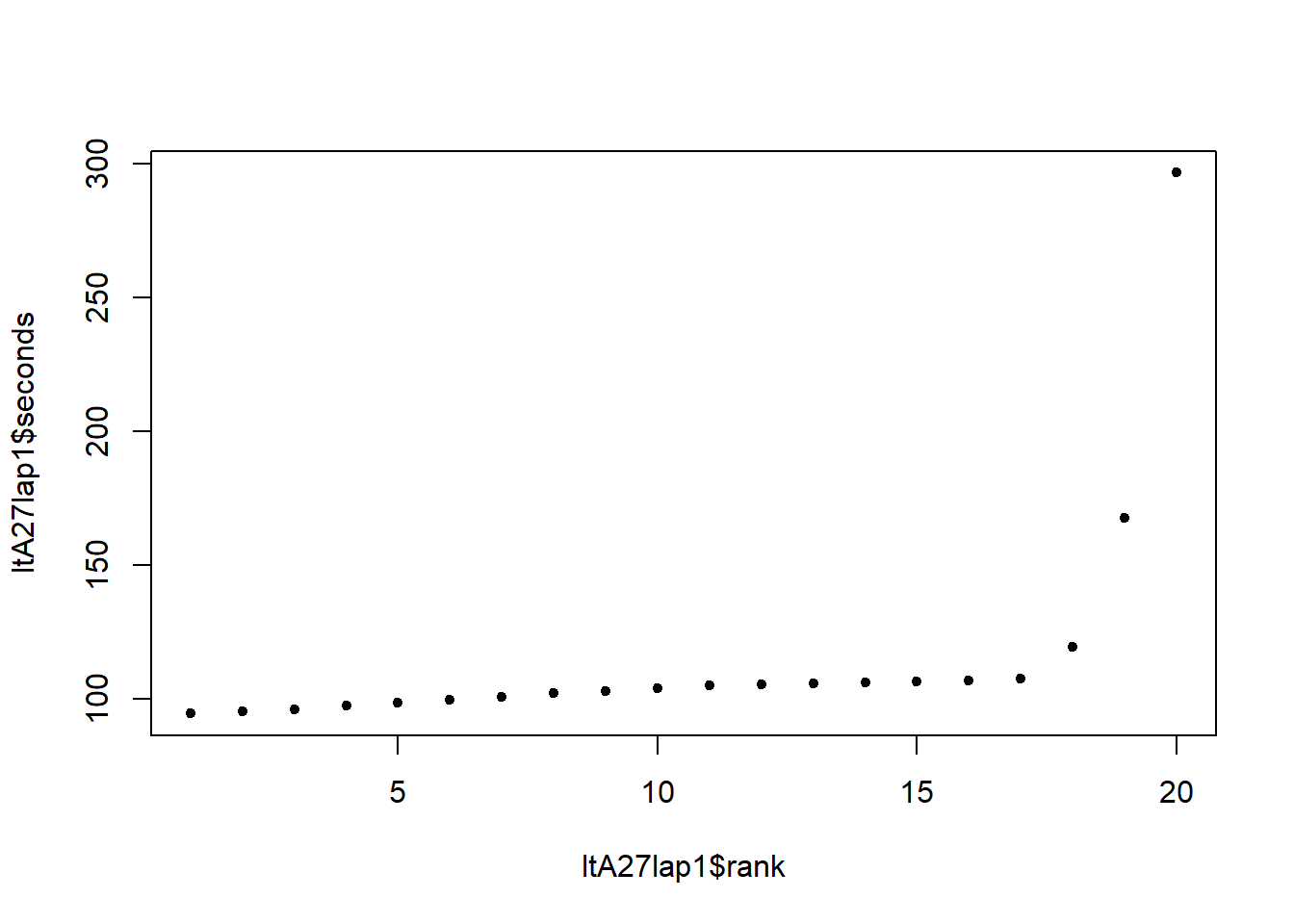
What happens if there are two identical lap times though? Should
these both get the same rank? Or be ranked next to each other in a
random order? frank does the latter by default. To give
them equal ranks, use the ties="dense" argument. I use this
all the time when I want to rank SNP positions to lay them out nicely on
a graphical genotype plot.
fcoalesce
Replaces missing values by stealing values from other columns. I’ve used it once, ever. I don’t know why I mentioned it here. Sorry.
The end
Well done! Like Lewis Hamilton in c(2008, 2014:2015, 2017:2020), your persistence has made you a world champion. Not at going around and around in a circle in a glorified go-kart for the benefit of gambling app developers, fossil fuel lobbies, and cigarette companies. No, you’re a world champion at data.table! Which is probably better for the world anyway.
